Tackling the Next Frontier in Single-Cell Sequencing
San Diego, June 11, 2013 -- You’ve heard of Mini-Me. Now along comes the uncomical field of Mini-Metagenomics. Building on their earlier successes in developing computational tools to assemble the genomes of individual bacterial cells, researchers have developed a method for sequencing the DNA of a collection of bacteria simultaneously – effectively decoding the genome of rare, low-abundance bacteria found on a hospital restroom sink.
|
“This bacteria was elusive and exotic, and it had not been sequenced,” said UC San Diego computer science and engineering professor Pavel Pevzner, a co-author of the paper* in the June 18 issue of the Proceedings of the National Academy of Sciences (PNAS), out this week. “This gives us hope that by building on our early innovations in single-cell assembly, we will be able to shine a light on even the least-known bacteria that were considered the ‘dark matter’ of life.”
Support for the research came from the Alfred P. Sloan Foundation, National Institutes of Health, and a Government of the Russian Federation Grant.
Approximately 1 percent of bacteria can be cultured and therefore easily sequenced. The other 99 percent have been called ‘dark matter’ because standard sequencing technologies typically require having at least one million of the cells on hand (an unlikely scenario, since few bacteria can be grown in the lab to a million cells). With the advent of single-cell assembly, however, scientists are sequencing these ‘dark’ bacteria one at a time – or in the latest case, many at a time.
“This is single-cell assembly on steroids,” observed Pevzner, an academic participant in Calit2's Qualcomm Institute at UC San Diego. “Think of it this way: traditional single-cell assembly is like assembling a puzzle with many lost and incorrect pieces. So assembling the mini-metagenome is like doing multiple puzzles simultaneously, all of which have lost and incorrect pieces. It’s a major computational task, and we proved that it can be done with the SPAdes assembler.”
|
Single-cell genomics is one of the hottest new genomics technologies, and much of the foundational research took place in San Diego – primarily in the JCVI lab of Roger Lasken, Pevzner’s lab in the Computer Science and Engineering department of UC San Diego’s Jacobs School of Engineering, and professor Glenn Tesler’s lab in UC San Diego’s Department of Mathematics. The assembly of mini-metagenomes was done using the SPAdes assembler, which was developed in the Algorithmic Biology Laboratory in Russia that Pevzner founded during a sabbatical in 2011.
While single-cell sequencing has primarily been used in marine and soil environments, the San Diego researchers’ PNAS paper is the second in as many months to focus on the sequencing of bacteria in places that may be of more immediate relevance to human health – namely, indoor environments.
In the May 2013 issue of the journal Genome Research**, Lasken, Pevzner and their colleagues published the genome of the pathogen Porphyromonas gingivalis. The journal’s cover showed a hospital sink against a stark white background. The pathogen was recovered from a biofilm in a hospital sink, and the researchers used a high-throughput, single-cell genomics platform to sequence it. For the first time, scientists proved that single-cell sequencing made it possible to take complex biofilm samples – in a hospital, for instance – and do a comparative genomic analysis of a pathogen’s strain variation.
|
As with the study published in Genome Research, the PNAS study specifically targeted bacterial cells within a complex biofilm from a hospital bathroom sink drain – indeed, the same bathroom sink.
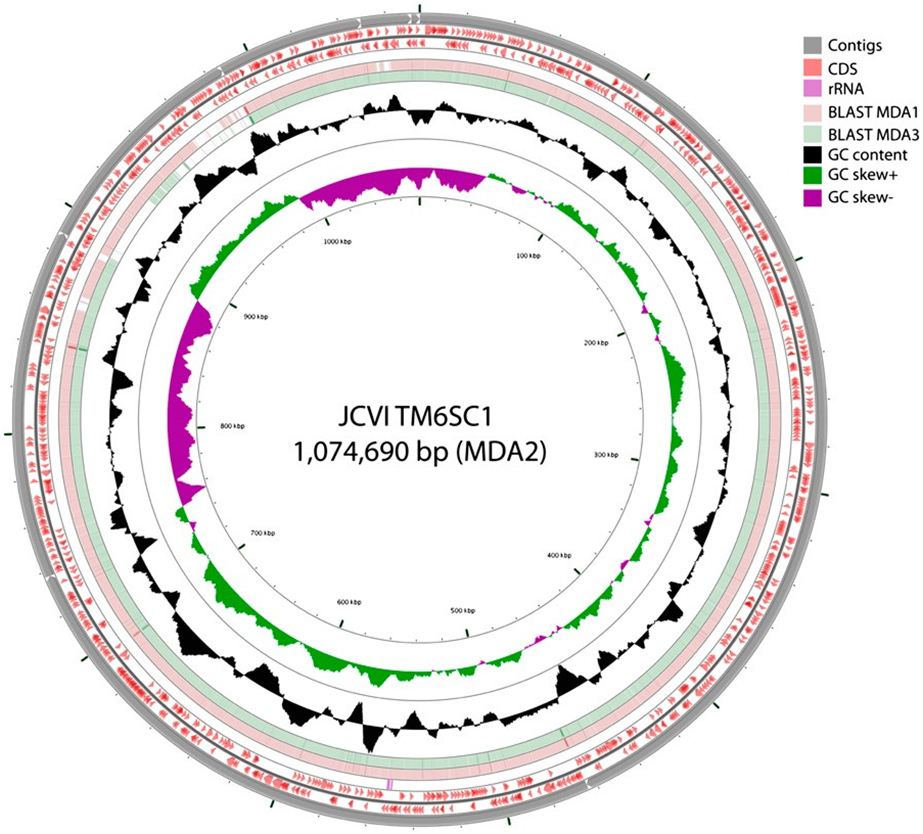
In addition to reconstructing more than 90 percent of a hitherto little-known phylum (officially designated TM6SC1), the PNAS work used the mini-metagenomic approach which includes high-throughput automation techniques and assembly tools to capture and sequence uncultivated bacterial species. In this case, it did so by amplifying and sequencing random pools of many cells simultaneously.
Biofilms are communities of bacteria often found on surfaces near water distribution systems such as sinks and shower heads. They are diverse microbial communities and could include disease-causing micro-organisms. Since these environments can serve as reservoirs of pathogenic bacteria, notably in hospitals and other health care facilities, there is considerable interest in understanding the abundant as well as rare bacterial species found in these biofilms.
And that’s the problem: Biofilms typically contain low overall cell numbers, and it is hard to separate rare bacteria from the organic and inorganic particulates present in the sample (which may emit the same fluorescent signal as the bacteria, so it’s hard to tell them apart).
|
Computational strategies were used to reconstruct and bin contigs representing genomes of individual species from the mixed genomes similar to published metagenomic methods, revealing a near-complete genome for TM6 (conservatively estimated at 91 percent completeness).
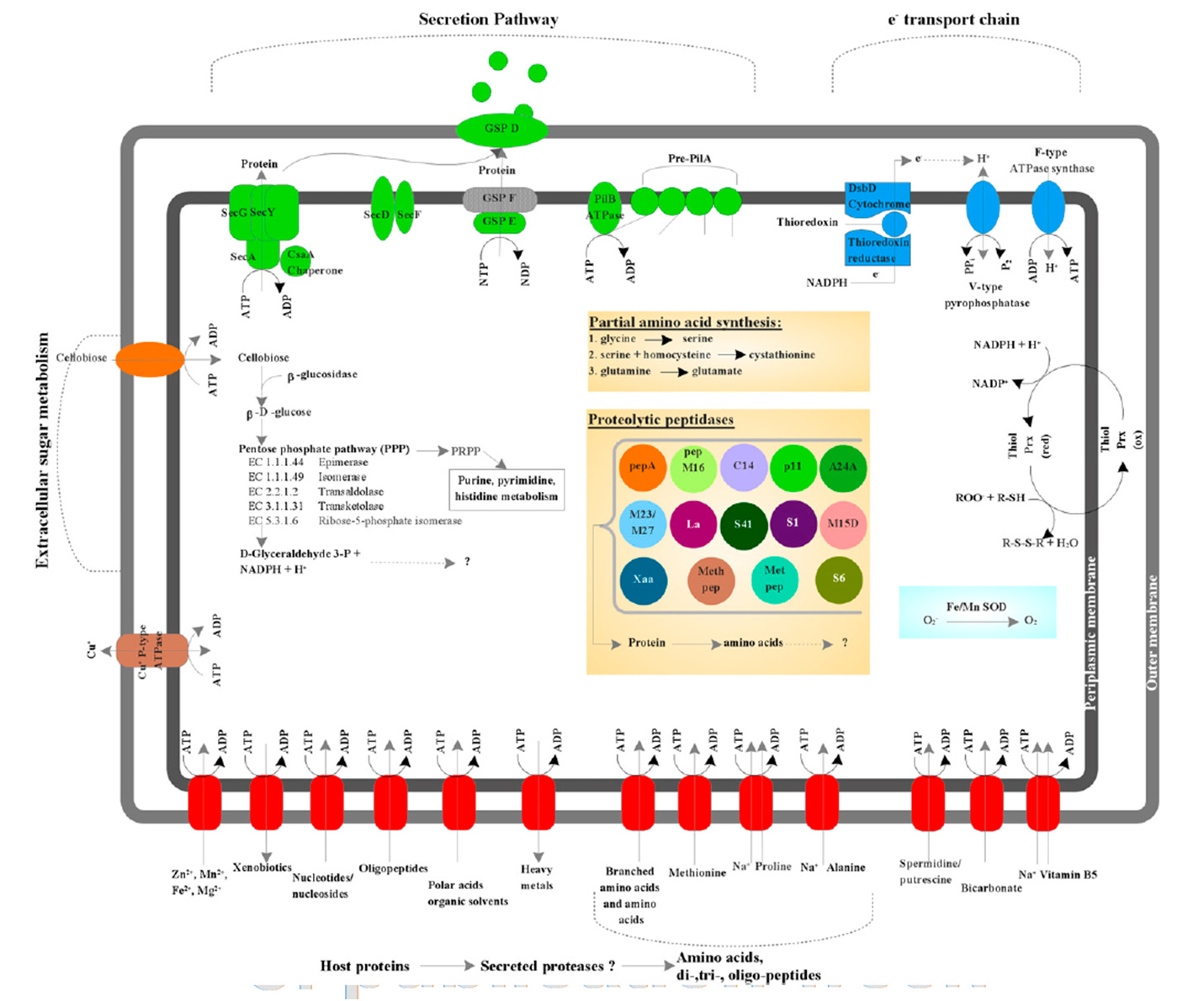
Previous studies identified TM6 using a phylogenetic marker (the 16S rRNA gene) in a number of diverse environments with global distribution. The most similar sequence on record is from a biofilm in a corroded copper water pipe. Based on the genomic information available, TM6 is likely a Gram-negative and facultatively anaerobic representative, with a low percentage of functionally annotated genes (43 percent). It can also generate energy from organic carbon sources with a modified electron transport chain adapted for both aerobic and anaerobic conditions. Based on the assembled genome, however, TM6 does not appear to have the capacity for de novo synthesis of amino acids.
What the genome did not resolve is whether TM6 is a free-living organism, or symbiont – forming a symbiotic relationship with an unknown host, possibly as an endosymbiont of an amoeba.
“We see a great opportunity to apply this approach to other environments to learn more about bacteria that have not been cultured to date,” said Pevzner. “This approach of single-cell sequencing applied to mini-metagenomics greatly increases the likelihood of capturing and assembling genomes of elusive micro-organisms that are not abundant. We can fill in gaps in the bacterial tree of life.”
Pevzner sees future applications of high-throughput, single-cell sequencing in the study of ‘microbial dark matter’ that may be useful in the development of new antibiotics. Along with his Russian collaborators, he is now moving from sequencing single bacterial cells to single cancer cells.
________________________________________
*Jeffrey S. McLean, et al., “Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum,” Proceedings of the National Academy of Sciences, July 2013. Doi: 10.1073/pnas.1219809110
**Jeffrey S. McLean, et al., “Genome of the pathogen fwewewewewe recovered from a biofilm in a hospital sink using a high-throughput single-cell genomics platform,” Genome Research, 23: 867-877, May 2013. Doi: 10.1101/gr.150433112
Media Contacts
Doug Ramsey, 858-822-5825, dramsey@ucsd.edu; Ioana Patringenaru, 858-822-0899, ipatrin@ucsd.edu; Heather Kowalski, 301-943-8879, hkowalski@jcvi.org
Related Links
Proceedings of the National Academy of Sciences
Genome Research Article
J. Craig Venter Institute
Researcher Sequence Dark Matter of Life
Jacobs School of Engineering
Computer Science and Engineering



.jpg)